Input Data
We have inferred a preliminary bone marrow-derived MØ (BMDM) regulatory network from various mouse strains stimulated with various agonists with stimulus duration ranging from 20 minutes to 48 hours. Transcriptional changes in the BMDMs were then assayed by gene expression microarray. We reduced the genes used for inferrence for this preliminary study to the top 4,000 highest expressing and most variant genes. Transcriptome profiling was integrated with roughly 14,139 functional gene associations from STRING and for each bicluster the genes’ promoter and 3’ UTR were fetched from UCSC genome browser and used for de novo discovery of cis-regulatory motifs via the MEME and Weeder motif discovery algorithms, respectively.
Global Properties
 We identified 267 biclusters with an average of 30 genes each (we note that each gene can fall in more than one bicluster). After filtering the biclusters to identify those with a high degree of co-expression (residual ≤ 0.30) and either a high quality upstream motif (e-value ≤ 0.05) or 3’ UTR motif (8bp or 7bp match to miRNA in miRBase), we identified 68 high-confidence biclusters. In total there are 95 cis-regulatory elements of which 11 are 3’ UTR cis-regulatory elements that match to known miRNAs in miRBase30. This preliminary modeling provides a large network describing the regulation of the 4,000 genes with most significant expression changes when BMDMs stimulated with various antigens.
We identified 267 biclusters with an average of 30 genes each (we note that each gene can fall in more than one bicluster). After filtering the biclusters to identify those with a high degree of co-expression (residual ≤ 0.30) and either a high quality upstream motif (e-value ≤ 0.05) or 3’ UTR motif (8bp or 7bp match to miRNA in miRBase), we identified 68 high-confidence biclusters. In total there are 95 cis-regulatory elements of which 11 are 3’ UTR cis-regulatory elements that match to known miRNAs in miRBase30. This preliminary modeling provides a large network describing the regulation of the 4,000 genes with most significant expression changes when BMDMs stimulated with various antigens.
Preliminary Biological Insights
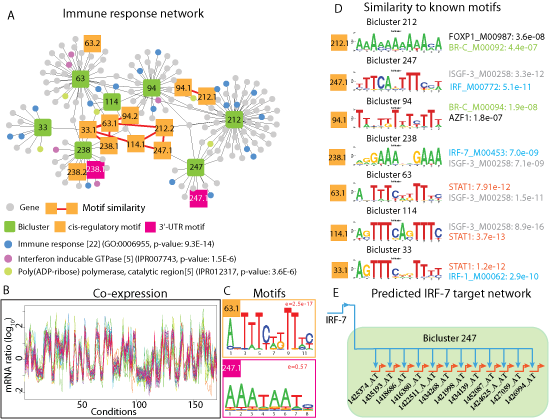
ISRE regulon of biclusters. A. Network diagram of regulon including genes, biclusters and regulatory motifs. B. Co-expression plot showing the expression of genes from a bicluster. C. Putative regulatory motifs discovered from cis-regulatory regions of the bicluster genes. D. Known TF-binding motifs similar to bicluster motifs. E. Irf7 regulates these genes through binding to the ISRE cis-regualtory element in from of these genes.
